”I am planning to diversify my research projects as part of my self-journey to exploration and exploitation of youth hood. Although I was a bit concentrated in Bioinformatics in high school, I found my hand in a physics lab at NYUAD where I basically do coding but under a different research context. I plan on being involved with Medical Imaging lab next semester in college.’

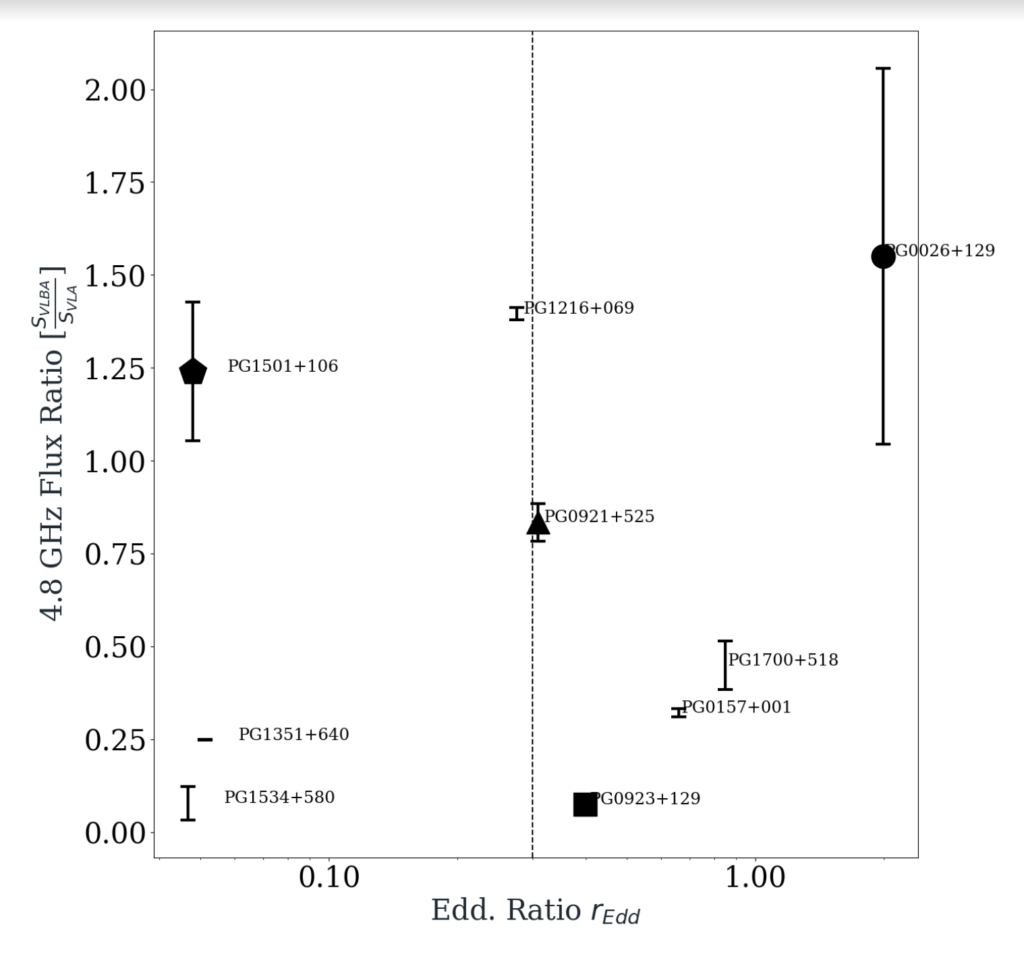
Dependence of Radio Emissions on the Eddington Ratio of supermassive black holes
Mentor: Dr. Ingyin Zaw (Program Head of Physics at NYUAD), Dr. Jospeph D Gelfand (Global Network Professor of Physics at New York University)
There are two classes of Black holes – black holes that emit radio emissions and black holes that don’t. In my research project, I am first using linux to inverse Fourier transform the image from 12 VLBA telescopes from all around the world to create a readable image of galaxies and its black hole. Then, I utilise several tools from CASA such as imfit and instate to study the properties of these generated images and classify black holes that emit radio emissions from those that don’t. I create an excel data sheet with all the info of the galaxies and use programming to perform statistical and visual analysis of the reduced AIPS data. After reduction in AIPS, I am basically acting as a data scientist. The tools that I use are pandas, matlplotlib, seaborn and lumpy packages from python.
Development of plant antibiotics against Seroheme CyCG Methyl Transferase using Computer Aided Drug Design (CADD)
Bacterial Wilt is one of the most prominent crop infections across villages in the Terai Region of Nepal. But Nepal does not have enough fertilizers and antibiotics to counter these malignant infections. I am currently working with one of Dr. Aryal’s students at Alpha Agro Pvt. Ltd. R&D Team to find an antibiotic against Bacterial Wilt. After complicated steps of protein prioritizations, we chose Seroheme Methyl Transferase as our protein target. Methyl Transferase (SAM) plays a critical role in the synthesis of Seroheme which is an integral co-factor in the sulfur assimilation pathway of bacteria. Inhibiting Seroheme would break the sulfur assimilation pathway and prevent respiration in bacteria. Our protein is a novel target. In-silico Molecular Docking, ADME analysis and other computational screening approaches were used in the project. With Dr. Aryal’s student, I predicted and induced mutation in our target protein using PYMOL and tested the efficacy of our lead compound.
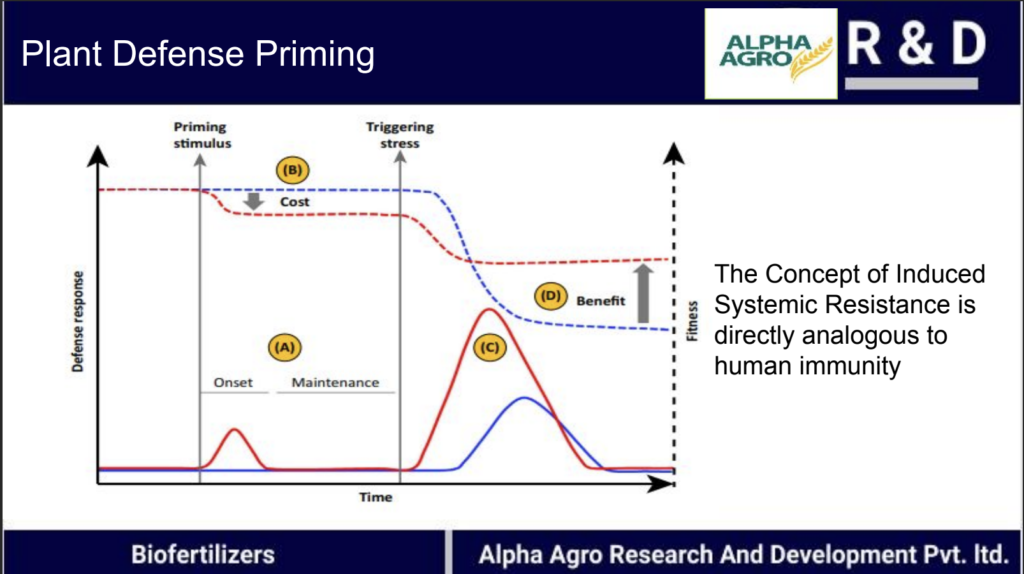
Exploiting plant ISR (Induced Systemic Resistance) and the evolutionary properties of the soil microbiome for disease prevention in tomato farming
As part of the need of Alpha Agro, I embarked on a journey to find possible molecules that could be used as plant vaccines to prevent plant infection. I found two molecules that could increase the Induced Systemic Resistance (ISR) of plants: Methyl Salicylate and Jasmonic acid. We are currently designing protocols for the organic synthesis of Methyl Salicylate and Jasmonic acid. Field Trial is on the way.
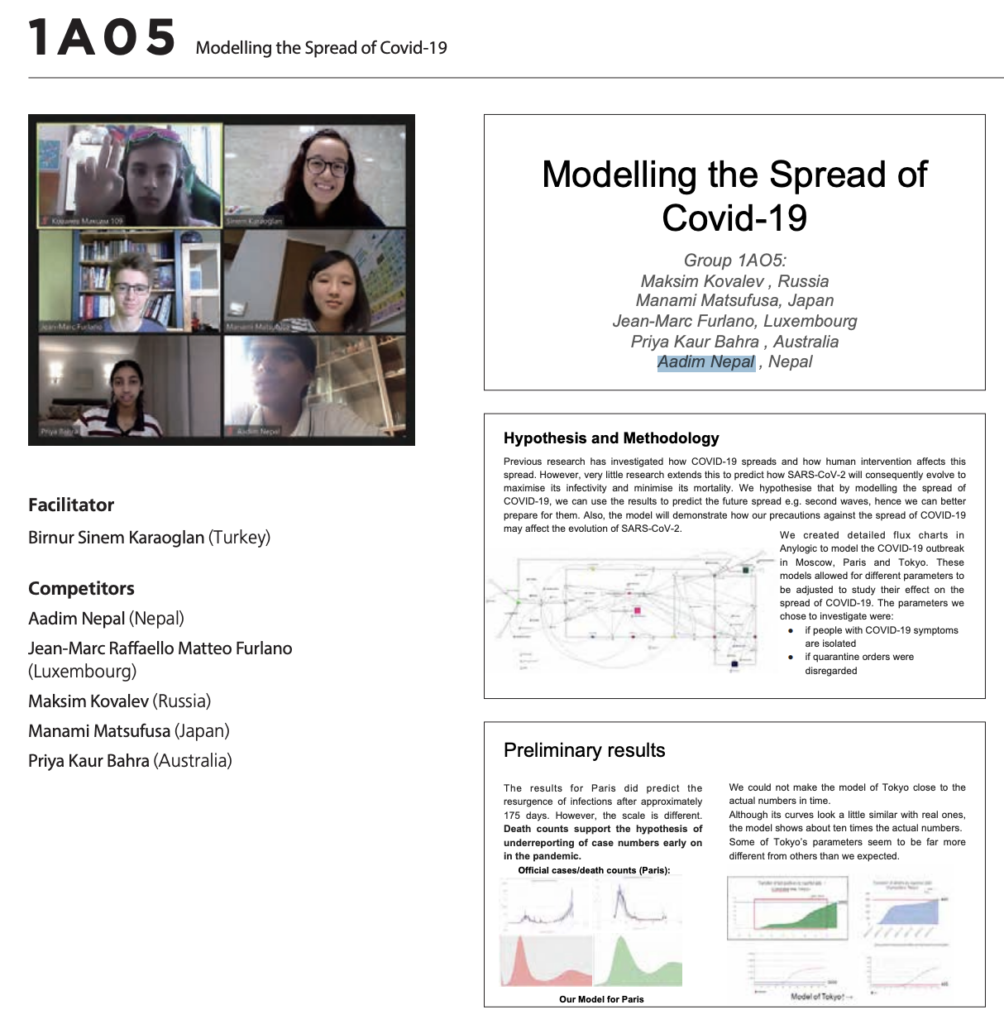
Epidemiological simulation of the evolution of SARS-CoV-2 in major cities including Paris and Moscow through models in “Any-Logic”
International Biology Olympiad Group Project (IBO 2020)
We created a detailed SIRS (Susceptible-Infectious-Recovered-Susceptible) flowchart as shown in the figure above in the software “Any-logic”. SIRS is one of the most practical epidemiological simulation models available in the world. After writing the whole model in anylogic, we derived all rates through variable parameters as follows:
rE1A = Exposed1(1-ProbInf)/(TotalIncPer-LateIncPer) rAR = Asymptomatic/AsympRecPer rE1E2 = Exposed1ProbInf/(TotalIncPer-LateIncPer)
rE2I = Exposed2/LateIncPer
rIL = Infectious/DetectPer
rLR = Light*(1-ProbLM)/LTransitionPerrLM = LightProbLM/LTransitionPer rMR = MediumProbMR/MTransitionPer
rMS = MediumProbMS/MTransitionPer rMD = Medium(1-ProbMS-ProbMR)/MTransitionPer
rSR = Severe(1-(TotalMortality-ProbLM+ProbLMProbMS+ProbLMProbMR)/(ProbLMPr
obMS))/STransitionPer
rSD = Severe((TotalMortality-ProbLM+ProbLMProbMS+ProbLMProbMR)/(ProbLMProb
MS))/STransitionPer
rRS = Recovered/ImmunLossPer
The parameters were obtained through a calibration experiment. COVID variables obtained from different government websites were entered in the model and thereby, the infection recurrence pattern prognosticated.
Designing Experiments for Target Validation of Novel Cyclin Dependent Kinase 12
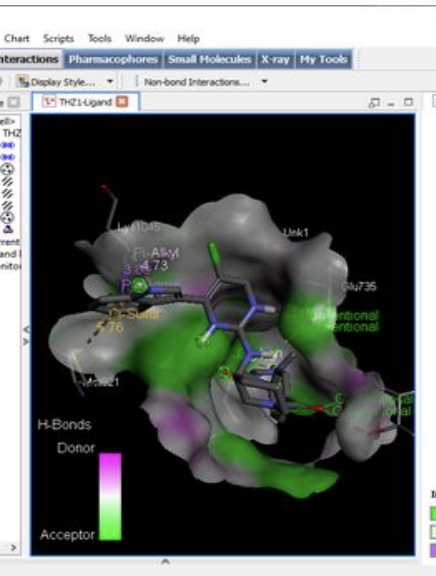
I took the lead in designing experiments (flow cytometry, western blot, IC50 Cell Viability) for target validation of novel Cyclin Dependent Kinase 12 (CDK12). CDK12 has a role in transcription. It phosphorylates the CTD terminal of RNA pol II which is an important transcription factor that helps encode DNA repairing enzymes. Target validation was the first part of the project. The second part included computational drug analysis (in-silico molecular docking). We proposed THZ to be the best drug candidate against CDK12 due to its excellent binding affinity score of -9.9 kcal/mol.
The Computational Biology part of the project was supervised by Dr. Pramod Aryal
Indole Derivative Indirubin as a possible drug scaffold against Cyclin Dependent Kinase 12 protein
Drugs like THZ have been proposed as a potential drug candidate against the novel CDK12. In my project, I propose Indirubin as a better drug candidate because of its property of strong inhibitive potential against all variants of CDK protein (i.e. CDK1, CDK5, CDK9, CDK12) that are a major player in cancer. In-silico molecular docking was used to obtain the inhibitive potential.
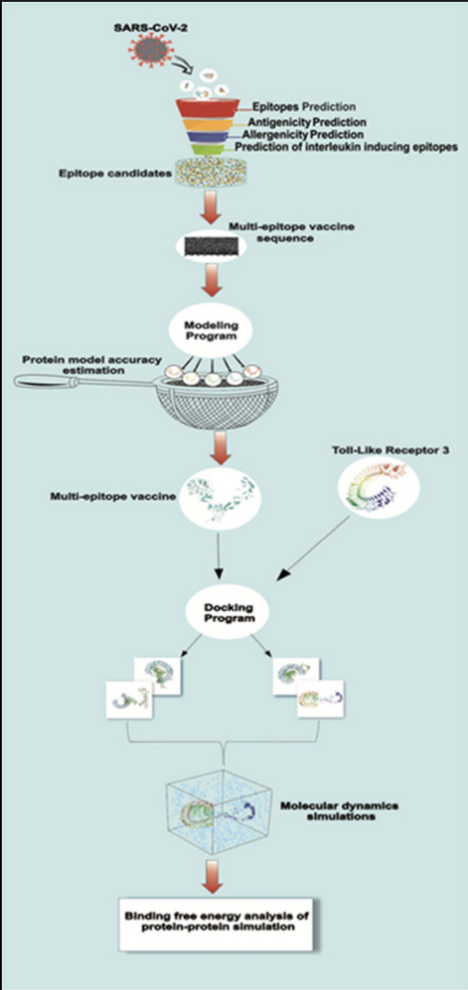
Implications of the use of live attenuated SARS-CoV-2 in vaccines
Research on the potential impacts of the use of attenuated SARS-CoV-2 in vaccines. My initial motivation to work on this background research project came after studying about “epitopes”. A single antigen contains multiple epitopes. So, it is intuitive to think that live SARS-CoV-2 might do good with vaccinations given the possible “multi-epitope” immune response. In my research, I highlight why “multi-epitope” response doesn’t necessarily entail a better immune response.